Fig. 7
From: MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton
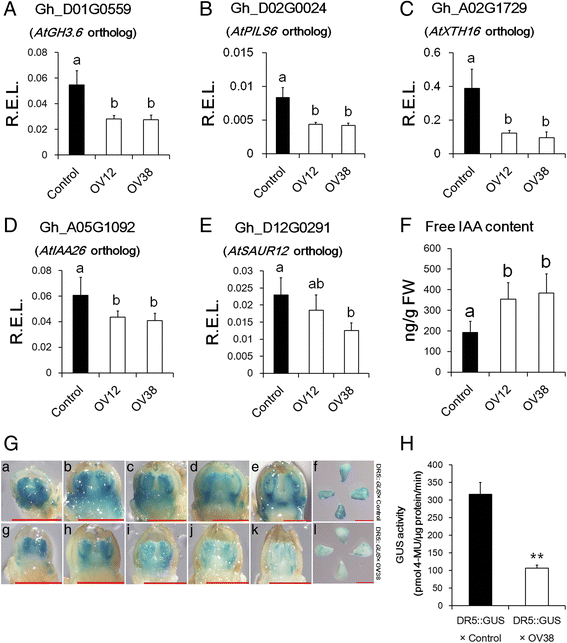
Auxin signalling was lower in over-expression lines than Control. a-e Real-time PCR analysis of auxin-inducible genes in floral buds. R.E.L., the relative expression levels calculated using HISTONE3 (AF024716) as a control. The error bars indicate the standard deviation of four biological replicates. Different letters indicate statistically significant differences at P < 0.05 based on analysis of variance (ANOVA) (Tukey’s multiple comparison test). f Quantitative analysis of IAA content in the floral buds of Control, OV12 and OV38 at −1 DPA. The error bars indicate the standard deviation of at least six biological replicates. Different letters indicate statistically significant differences at P < 0.05 based on analysis of variance (ANOVA) (Tukey’s multiple comparison test). g GUS staining images of floral buds and ovules. Images of floral buds (length ≤ 2 mm) from DR5::GUS × Control (a-e) and DR5::GUS × OV38 (g-k) and ovules from DR5::GUS × Control (f) and DR5::GUS × OV38 (l) at −1 DPA. Scale bar, 1 mm. h Quantitative GUS assays of ovules at −1 DPA. Asterisks indicate statistically significant differences at P < 0.01 based on Student’s t test. Control, nontransgenic plant segregated from 35S::GhmiR157 transgenic lines in cotton. OV12 and OV38, 35S::GhmiR157 transgenic lines in cotton. DR5::GUS × Control and DR5::GUS × OV38 were F1 plants of DR5::GUS crossed with Control and OV38, respectively